Microbial Ecological Diversity Characteristics of the Soil Profile in the Vadose Zone Polluted by Ammonia Nitrogen
-
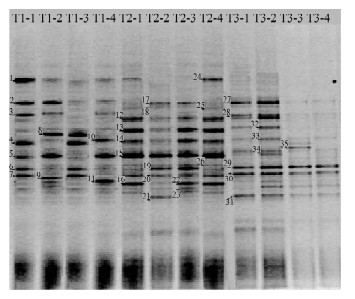
摘要: 包气带土壤中氨氮污染迁移规律已成为近年来国内外学者研究的重点,目前在氨氮污染迁移规律研究中采用柱实验模拟以及软件模拟的方法较多,土壤生物技术则广泛应用于污染物降解领域,而在污染物迁移规律研究中应用较少。本文采用变性梯度凝胶电泳(DGGE)技术、16S rRNA序列分析技术以及典范对应分析相结合,对华北平原3个典型氨氮污染区土壤表层至包气带剖面微环境的细菌垂直分布特征及群落结构进行研究。结合污染区土壤理化性质分析,认为包气带土壤剖面的细菌群落中存在着与氮循环、硫酸盐代谢等过程偶联的优势细菌类群,说明土壤微环境中细菌群落分布明显受氨态氮、硝态氮、亚硝态氮和硫酸盐的分布影响,进一步表明污染土壤优势菌群的群落结构信息是描述包气带土壤环境氨氮污染物迁移规律的重要参数。Abstract: Scholars at home and abroad in recent years have focused on researching ammonia nitrogen pollution migration rules in the vadose zone soil. Column simulation and software simulation were primarily conducted for the study of ammonia nitrogen pollution migration. Biotechnology was widely used in degradation of pollutant in soil, but less applied to the study of pollution migration. This study applied Denaturing Gradient Gel Electrophoresis (DGGE) and the sequence analysis of the V3 Region of 16S rRNA combined with canonical correspondence analysis to characterize the bacteria vertical distribution characteristics and bacterial community structure in the soil from three typical contaminated zones in the North China Plain. According to the analysis of physical and chemical soil properties in polluted areas, there were some dominant individual bacteria in the key pathways of the nitrogen cycle and sulfate metabolism. It is suggested that the bacterial communities are affected by the distributions of ammonia, nitrate and nitrite nitrogen, showing that the community structure information of the dominant population in contaminated soil is an important parameter to study the ammonia nitrogen pollution migration rules.
-
-
表 1 土壤样品环境因子测定方法
Table 1. Determination of environmental factors in soil samples
测试项目 测试方法 土壤含水量 质量法 pH值 电化学分析法 氧化还原电位 电位法 颗粒组成 比重计法 铵态氮 纳氏比色法 硝态氮、总氮 紫外分光光度法 亚硝态氮 N-(1-萘基)-
乙二胺光度法阳离子 电感耦合等离子体
发射光谱法其他阴离子 离子色谱法 表 2 引物序列
Table 2. Primer sequence
引物 序列 341F 5′-CCTACGGGAGGCAGCAG-3′ 534R 5′-ATTACCGCGGCTGCTGG-3′ 341F-GC 5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGG
GGGCACGGGGGGCCTACGGGAGGCAGCAG-3′M13-47 5′-TAATACGACTCACTATAGGGC-3′ RV-M 5′-ATTTAGGTGACACTATAGAAT ACTC-3′ 表 3 各点位土壤样品理化参数及微生物数量统计
Table 3. physicochemical factors and microorganisms of soil samples
站点 土壤岩性 细菌总数/
(CUF·g-1)深度/m 酸碱度
pH含水率/% 氧化还原电位
Eh/mV干重wB/(mg·kg-1) NH4-N NO3-N NO2-N SO42- T1-1 粉质黏土 5.37×109 1.94 8.73 20.53 247.00 2.24 0.46 0.08 56.22 T1-2 黏土夹砂 7.27×108 3.27 8.73 16.75 201.00 0.52 0.47 0.12 36.61 T1-3 粉细砂 8.17×106 7.45 8.63 18.34 133.10 0.42 0.69 0.13 9.82 T1-4 细砂 2.64×105 8.21 8.68 17.26 107.20 0.53 1.96 0.12 6.62 T2-1 黏土 8.87×109 0.95 8.14 17.77 566.00 21.04 22.21 16.88 19.04 T2-2 粉土 1.53×108 2.21 8.71 17.75 187.60 4.88 5.09 0.35 24.38 T2-3 粉细砂 8.31×106 4.00 8.33 3.83 284.00 9.97 2.04 0.43 49.83 T2-4 粉土 9.53×106 8.15 8.56 9.83 226.00 66.59 11.61 0.81 33.29 T3-1 黏土夹砂 3.22×107 0.27 8.55 0.17 491.00 0.46 4.56 1.34 448.05 T3-2 粉土含砂 1.54×108 1.23 8.63 19.17 372.00 1.11 4.06 2.47 29.10 T3-3 中细砂 8.00×106 3.25 8.62 18.02 90.70 2.48 3.21 0.59 45.75 T3-4 粗砂 1.60×105 4.21 8.73 16.87 86.00 5.16 3.68 0.52 28.00 -
[1] William J H. Vadose zone microbial biobarriers remove nitrate from percolating groundwater [J]. Current Microbiology, 2009(58): 622-627.
[2] Mamie N I, Kate M S, Dennis E R. Reduction of perchlorate and nitrate by microbial communities in vadose soil [J]. Applied and Environmental Micro-biology, 2005, 71(7): 3928-3934. doi: 10.1128/AEM.71.7.3928-3934.2005
[3] 卞华松,张仲燕.冷冻固定化优势菌群处理含甲醛苯酚废水[J].环境科学,1998,19(2): 39-42. http://www.cnki.com.cn/Article/CJFDTOTAL-HJKZ802.009.htm
[4] 赵娟,吕剑,何义亮,靳强,张文英,何霞.异养脱氮菌株Bacillus sp. LY 降解有毒有机污染物的研究[J].环境科学,2007,28(12): 2838-2842. doi: 10.3321/j.issn:0250-3301.2007.12.030
[5] 袁勇军,陆兆新,黄丽金,吕凤霞,别小妹.烟碱降解细菌的分离鉴定及其降解性能的初步研究[J].微生物学报,2005,45(2): 181-184. http://www.cnki.com.cn/Article/CJFDTOTAL-WSXB200502005.htm
[6] 洪青,张忠辉,张晓舟,徐剑宏,李顺鹏.中度嗜盐菌Halomonas sp. BYS2 1启动子的克隆和测序[J].应用与环境生物学报,2005,11(6): 729-732.
[7] Myers R M, Fischer S G, Maniatis T. Modification of the melting properties of duplex DNA by attachment of a GC-rich DNA sequence as determined by denaturing gradient gel electrophoresis [J]. Nucleic Acids Research, 1985, 13: 3111-3129. doi: 10.1093/nar/13.9.3111
[8] Muyzer G, Brinkhoff T, Nübel U. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology [J]. Molecular Microbial Ecology Manual,1998,3(44): 1-27.
[9] Zweifel U L, Hagstrom A. Total counts of marine bacteria include a large fraction of non- nucleoid-containing bacteria (ghosts) [J].Applied and Environ-mental Microbiology, 1995, 61(6): 2180-2185.
[10] Schallenberg M, Kalff J, Rasmussen J B. Solutions to problems in enumerating sediment bacteria by direct count[J].Applied and Environmental Microbiology, 1989, 55(5): 1214-1219.
[11] Zhou J Z, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition [J]. Applied and Environ-mental Microbiology, 1996, 62(2): 316-322.
[12] Muyzer G, Dewaal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA[J]. Applied and Environmental Microbiology, 1993(59): 695-700.
[13] Rohlf F J. NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System, Version 2.0 [M].New York: State University of New York, 2000: 97.
[14] Thompson J D, Higgins D G, Gibson T J, Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice[J]. Nucleic Acids Research, 1994(22): 4673-4680.
[15] Kumar S, Tamura K, Nei M. MEGA 3: Integrated software for molecular evolutionary genetics analysis and sequence alignment [J]. Briefings in Bioinformatics, 2004(5): 150-163.
[16] Schloss P D, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness [J]. Applied and Environmental Microbiology, 2005(71): 1501-1506.
[17] Selim S, Negrel J, Govaerts C, Gianinazzi S, van Tuinen D.Isolation and partial characterization of antagonistic peptides produced by Paenibacillus sp. Strain B2 isolated from the Sorghum Mycorrhizosphere[J]. Applied and Environmental Microbiology, 2005(71): 6501-6507.
[18] Lin Y, Kong H N, He Y L. Simultaneous nitrification and denitrification in a membrane bioreactor and isolation of heterotrophic nitrifying bacteria [J]. Japanese Journal of Water Treatment Biology, 2004, 40(3): 105-114. doi: 10.2521/jswtb.40.105
[19] Derek R L, Elizabeth J P P. Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria [J]. Applied and Environ-mental Microbiology, 1994(60): 2394-2399.
[20] Robertson L A, Kuenen J G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, faculta-tively autotrophic sulphur bacterium [J]. General Microbiology, 1983, 129(9): 2847-2855.
[21] Gupta A B. Thiosphaera pantotropha: A sulphur bacterium capable of simultaneous heterotrophic nitrification and aerobic denitrification[J]. Enzyme & Microbial Technology, 1997(21): 589-595.
[22] Moir J W B, Wehrfritz J M, Spiro S. The biochemical characterization of a novel non-haem-iron hydroxylamine oxidase from Paracoccus denitrificans GB17 [J]. Biochemical Journal, 1996, 319(3): 823-827. doi: 10.1042/bj3190823
[23] Stephen P, Cummings D, Gilmour J. The effect of NaCl on the growth of a halomonas species: Accumulation and utilization of compatible solutes [J]. Microbiology, 1995(141): 1413-1418.
[24] Kim H, Bram V, Lieven W, Willy V, Nico B, Paul D V.Cultivation of denitrifying bacteria: Optimization of isolation conditions and diversity study [J]. Applied and Environmental Microbiology, 2006(72): 2637-2643.
[25] Stefan J G, Om P, Thomas M G. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination [J]. Applied and Environ-mental Microbiology, 2010(76): 3244-3254.
[26] Christiane W, Andrea T, Antje W. Horizon-specific bacterial community composition of german grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes [J]. Applied and Environmental Microbiology, 2010(76): 6751-6759.
[27] Nicole D, Bruno G, Steffen K. Methanotrophic communities in Brazilian Ferralsols from naturally forested, afforested, and agricultural sites[J]. Applied and Environmental Microbiology, 2010(76): 1307-1310.
-


 下载:
下载: